CEGR research is aimed at understanding how eukaryotic genes are regulated at a molecular level. Our physical location within shared space maximizes interactions that promote innovative thinking. Each lab brings unique strengths that together achieve a level of synergy that is rarely matched in scientific research. This includes excellence in biochemical assembly mechanisms in chromatin and transcription. It includes methods X-ray crystallography, cryo-EM, and a variety of biophysical methods to elucidate the structure and mechanism by which proteins interact with nucleosomes. We aim to understand how the bizarre repetitive structure of RNA polymerase II C-terminal domain regulates the transcription cycle. As we all know, specificity in gene regulation boils down to proteins selectively recognizing specific features (sequence and shape) on DNA, as allowed by the chromatin landscape, then using these anchor points to coelsce transcription complex assembly. We have developed state-of-the-art concepts about how DNA binding proteins access DNA that is encased in chromatin. We are studying how this gives rise to cell-type specific gene expression, for example, in making motor neurons or red blood cells. Through our experiments on human and mouse model systems we develop concepts about transcriptional regulation goes awry in cancer, and how we might use this knowledge to develop rectifying therapeutic drugs.
Publications
2023
| Systematic Dissection of Sequence Features Affecting the Binding Specificity of a Pioneer Factor Reveals Binding Synergy Between FOXA1 and AP-1. Xu C, Kleinschmidt H, Yang J, Leith E, Johnson J, Tan S, Mahony S, Bai L. bioRxiv. 2023 Nov 11:2023.11.08.566246. | |
| SEM: sized-based expectation maximization for characterizing nucleosome positions and subtypes. Yang J, Yen K, Mahony S. bioRxiv. 2023 Oct 20:2023.10.17.562727. | |
| Advances in direct detection of lysine methylation and acetylation by nuclear magnetic resonance using 13C-enriched cofactors. Fraser OA, Namitz KEW, Showalter SA. Methods. 2023 Oct;218:72-83. | |
| Allo: Accurate allocation of multi-mapped reads enables regulatory element analysis at repeats. Morrissey A, Shi J, James DQ, Mahony S. bioRxiv. 2023 Sep 15:2023.09.12.556916. | |
| Assessment of the roles of Spt5-nucleic acid contacts in promoter proximal pausing of RNA polymerase II. Dollinger R, Deng EB, Schultz J, Wu S, Deorio HR, Gilmour DS. J Biol Chem. 2023 Sep;299(9):105106. | |
| New roles for elongation factors in RNA polymerase II ubiquitylation and degradation. Reese JC. Biochim Biophys Acta Gene Regul Mech. 2023 Sep;1866(3):194956. | |
| Using Synthetic DNA Libraries to Investigate Chromatin and Gene Regulation. Kleinschmidt H, Xu C, Bai L. Chromosoma. 2023 Sep;132(3):167-189. | |
| Catalytic and non-catalytic mechanisms of histone H4 lysine 20 methyltransferase SUV420H1. Abini-Agbomson S, Gretarsson K, Shih RM, Hsieh L, Lou T, De Ioannes P, Vasilyev N, Lee R, Wang M, Simon MD, Armache JP, Nudler E, Narlikar G, Liu S, Lu C, Armache KJ. Mol Cell. 2023 Aug 17;83(16):2872-2883.e7. | |
| Dual engagement of the nucleosomal acidic patches is essential for deposition of histone H2A.Z by SWR1C. Baier AS, Gioacchini N, Eek P, Tan S, Peterson CL. Res Sq. 2023 Jul 28:rs.3.rs-3050911. | |
| Comparative proteomics of vesicles essential for the egress of Plasmodium falciparum gametocytes from red blood cells. Sassmannshausen J, Bennink S, Distler U, Küchenhoff J, Minns AM, Lindner SE, Burda PC, Tenzer S, Gilberger TW, Pradel G. Mol Microbiol. 2023 Jul 26. | |
| Long-read genome assembly and gene model annotations for the rodent malaria parasite Plasmodium yoelii 17XNL. Godin MJ, Sebastian A, Albert I, Lindner SE. J Biol Chem. 2023 Jul;299(7):104871. | |
| Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking. Chio US, Palovcak E, Autzen AAA, Autzen HE, Muñoz EN, Yu Z, Wang F, Agard DA, Armache JP, Narlikar GJ, Cheng Y. bioRxiv. 2023 Jun 20:2023.06.20.545796. | |
| RETROFIT: Reference-free deconvolution of cell-type mixtures in spatial transcriptomics. Singh R, He X, Park AK, Hardison RC, Zhu X, Li Q. bioRxiv. 2023 Jun 9:2023.06.07.544126. | |
| Investigating pioneer factor activity and its coordination with chromatin remodelers using integrated synthetic oligo assay. Chen H, Yan C, Dhasarathy A, Kladde M, Bai L. STAR Protoc. 2023 Jun 7;4(2):102279. | |
| Molecular basis of polycomb group protein-mediated fetal hemoglobin repression. Qin K, Lan X, Huang P, Saari MS, Khandros E, Keller CA, Giardine B, Abdulmalik O, Shi J, Hardison RC, Blobel GA. Blood. 2023 Jun 1;141(22):2756-2770. | |
| SARS-CoV-2 polyprotein substrate regulates the stepwise Mpro cleavage reaction. Narwal M, Armache JP, Edwards TJ, Murakami KS. J Biol Chem. 2023 May;299(5):104697. | |
| The mitochondrion of Plasmodium falciparum is required for cellular acetyl-CoA metabolism and protein acetylation. Nair SC, Munro JT, Mann A, Llinás M, Prigge ST. Proc Natl Acad Sci U S A. 2023 Apr 25;120(17):e2210929120. | |
| Basic helix-loop-helix pioneer factors interact with the histone octamer to invade nucleosomes and generate nucleosome-depleted regions. Donovan BT, Chen H, Eek P, Meng Z, Jipa C, Tan S, Bai L, Poirier MG. Mol Cell. 2023 Apr 20;83(8):1251-1263.e6. | |
| Cryo-EM structure of the human Sirtuin 6-nucleosome complex. Chio US, Rechiche O, Bryll AR, Zhu J, Leith EM, Feldman JL, Peterson CL, Tan S, Armache JP. Sci Adv. 2023 Apr 14;9(15):eadf7586. | |
| Ribosome heterogeneity and specialization of Plasmodium parasites. McGee JP, Armache JP, Lindner SE. PLoS Pathog. 2023 Apr 13;19(4):e1011267. | |
| Snapshot: a package for clustering and visualizing epigenetic history during cell differentiation. Xiang G, Giardine B, An L, Sun C, Keller CA, Heuston EF, Anderson SM, Kirby M, Bodine D, Zhang Y, Hardison RC. BMC Bioinformatics. 2023 Mar 20;24(1):102. | |
| Catalytic and non-catalytic mechanisms of histone H4 lysine 20 methyltransferase SUV420H1. Abini-Agbomson S, Gretarsson K, Shih RM, Hsieh L, Lou T, De Ioannes P, Vasilyev N, Lee R, Wang M, Simon M, Armache JP, Nudler E, Narlikar G, Liu S, Lu C, Armache KJ. bioRxiv. 2023 Mar 19:2023.03.17.533220. | |
| Cryo-EM structure of the human Sirtuin 6-nucleosome complex. Chio US, Rechiche O, Bryll AR, Zhu J, Feldman JL, Peterson CL, Tan S, Armache JP. bioRxiv. 2023 Mar 18:2023.03.17.533206. | |
| Predicting nucleosome positioning using statistical equilibrium models in budding yeast. Kharerin H, Bai L. STAR Protoc. 2023 Mar 17;4(1):101926. | |
| Cripowellins Pause Plasmodium falciparum Intraerythrocytic Development at the Ring Stage. Butler JH, Painter HJ, Bremers EK, Krai P, Llinás M, Cassera MB. Molecules. 2023 Mar 13;28(6):2600. | |
| Cytoplasmic isoleucyl tRNA synthetase as an attractive multistage antimalarial drug target. Istvan ES, Guerra F, Abraham M, Huang KS, Rocamora F, Zhao H, Xu L, Pasaje C, Kumpornsin K, Luth MR, Cui H, Yang T, Palomo Diaz S, Gomez-Lorenzo MG, Qahash T, Mittal N, Ottilie S, Niles J, Lee MCS, Llinas M, Kato N, Okombo J, Fidock DA, Schimmel P, Gamo FJ, Goldberg DE, Winzeler EA. Sci Transl Med. 2023 Mar 8;15(686):eadc9249. | |
| Gene silencing dynamics are modulated by transiently active regulatory elements. Vermunt MW, Luan J, Zhang Z, Thrasher AJ, Huang A, Saari MS, Khandros E, Beagrie RA, Zhang S, Vemulamada P, Brilleman M, Lee K, Yano JA, Giardine BM, Keller CA, Hardison RC, Blobel GA. Mol Cell. 2023 Mar 2;83(5):715-730.e6. | |
| A Copper-Responsive Two-Component System Governs Lipoprotein Remodeling in Listeria monocytogenes. Komazin G, Rizk AA, Armbruster KM, Bonnell VA, Llinás M, Meredith TC. J Bacteriol. 2023 Jan 26;205(1):e0039022. | |
| Long-Read Genome Assembly and Gene Model Annotations for the Rodent Malaria Parasite Plasmodium yoelii 17XNL. Godin MJ, Sebastian A, Albert I, Lindner SE. bioRxiv. 2023 Jan 7:2023.01.06.523040. | |
| A direct nuclear magnetic resonance method to investigate lysine acetylation of intrinsically disordered proteins. Fraser OA, Dewing SM, Usher ET, George C, Showalter SA. Front Mol Biosci. 2023 Jan 6;9:1074743. |
2022
| CLIMB: High-dimensional association detection in large scale genomic data. Koch H, Keller CA, Xiang G, Giardine B, Zhang F, Wang Y, Hardison RC, Li Q. Nat Commun. 2022 Nov 12;13(1):6874. | |
| CTCF blocks antisense transcription initiation at divergent promoters. Luan J, Vermunt MW, Syrett CM, Coté A, Tome JM, Zhang H, Huang A, Luppino JM, Keller CA, Giardine BM, Zhang S, Dunagin MC, Zhang Z, Joyce EF, Lis JT, Raj A, Hardison RC, Blobel GA. Nat Struct Mol Biol. 2022 Nov;29(11):1136-1144. | |
| Intranasal SARS-CoV-2 RBD decorated nanoparticle vaccine enhances viral clearance in the Syrian hamster model. Patel DR, Minns AM, Sim DG, Field CJ, Kerr AE, Heinly T, Luley EH, Rossi RM, Bator C, Moustafa IM, Hafenstein SL, Lindner SE, Sutton TC. bioRxiv. 2022 Dec 14:2022.10.27.514054. | |
| Piperaquine-resistant PfCRT mutations differentially impact drug transport, hemoglobin catabolism and parasite physiology in Plasmodium falciparum asexual blood stages. Okombo J, Mok S, Qahash T, Yeo T, Bath J, Orchard LM, Owens E, Koo I, Albert I, Llinás M, Fidock DA. PLoS Pathog. 2022 Oct 28;18(10):e1010926. | |
| Biophysical insights into glucose-dependent transcriptional regulation by PDX1. Usher ET, Showalter SA. J Biol Chem. 2022 Dec;298(12):102623. | |
| The anticancer human mTOR inhibitor sapanisertib potently inhibits multiple Plasmodium kinases and life cycle stages. Arendse LB, Murithi JM, Qahash T, Pasaje CFA, Godoy LC, Dey S, Gibhard L, Ghidelli-Disse S, Drewes G, Bantscheff M, Lafuente-Monasterio MJ, Fienberg S, Wambua L, Gachuhi S, Coertzen D, van der Watt M, Reader J, Aswat AS, Erlank E, Venter N, Mittal N, Luth MR, Ottilie S, Winzeler EA, Koekemoer LL, Birkholtz LM, Niles JC, Llinás M, Fidock DA, Chibale K. Sci Transl Med. 2022 Oct 19;14(667):eabo7219. | |
| Inhibitors of ApiAP2 protein DNA binding exhibit multistage activity against Plasmodium parasites. Russell TJ, De Silva EK, Crowley VM, Shaw-Saliba K, Dube N, Josling G, Pasaje CFA, Kouskoumvekaki I, Panagiotou G, Niles JC, Jacobs-Lorena M, Denise Okafor C, Gamo FJ, Llinás M. PLoS Pathog. 2022 Oct 12;18(10):e1010887. | |
| Origin recognition complex harbors an intrinsic nucleosome remodeling activity. Li S, Wasserman MR, Yurieva O, Bai L, O'Donnell ME, Liu S. Proc Natl Acad Sci U S A. 2022 Oct 18;119(42):e2211568119. | |
| Partitioned usage of chromatin remodelers by nucleosome-displacing factors. Chen H, Kharerin H, Dhasarathy A, Kladde M, Bai L. Cell Rep. 2022 Aug 23;40(8):111250. | |
| Nucleosome-directed replication origin licensing independent of a consensus DNA sequence. Li S, Wasserman MR, Yurieva O, Bai L, O'Donnell ME, Liu S. Nat Commun. 2022 Aug 23;13(1):4947. | |
| Understanding the Excitation Wavelength Dependence and Thermal Stability of the SARS-CoV-2 Receptor-Binding Domain Using Surface-Enhanced Raman Scattering and Machine Learning. Zhang K, Wang Z, Liu H, Perea-López N, Ranasinghe JC, Bepete G, Minns AM, Rossi RM, Lindner SE, Huang SX, Terrones M, Huang S. ACS Photonics. 2022 Aug 25;9(9):2963-2972. | |
| Mitochondrially targeted proximity biotinylation and proteomic analysis in Plasmodium falciparum. Lamb IM, Rios KT, Shukla A, Ahiya AI, Morrisey J, Mell JC, Lindner SE, Mather MW, Vaidya AB. PLoS One. 2022 Aug 19;17(8):e0273357. | |
| Identification of basal complex protein that is essential for maturation of transmission-stage malaria parasites. Clements RL, Morano AA, Navarro FM, McGee JP, Du EW, Streva VA, Lindner SE, Dvorin JD. Proc Natl Acad Sci U S A. 2022 Aug 23;119(34):e2204167119. | |
| HIC2 controls developmental hemoglobin switching by repressing BCL11A transcription. Huang P, Peslak SA, Ren R, Khandros E, Qin K, Keller CA, Giardine B, Bell HW, Lan X, Sharma M, Horton JR, Abdulmalik O, Chou ST, Shi J, Crossley M, Hardison RC, Cheng X, Blobel GA. Nat Genet. 2022 Sep;54(9):1417-1426. | |
| Development and Validation of Indirect Enzyme-Linked Immunosorbent Assays for Detecting Antibodies to SARS-CoV-2 in Cattle, Swine, and Chicken. Gontu A, Marlin EA, Ramasamy S, Neerukonda S, Anil G, Morgan J, Quraishi M, Chen C, Boorla VS, Nissly RH, Jakka P, Chothe SK, Ravichandran A, Kodali N, Amirthalingam S, LaBella L, Kelly K, Natesan P, Minns AM, Rossi RM, Werner JR, Hovingh E, Lindner SE, Tewari D, Kapur V, Vandegrift KJ, Maranas CD, Surendran Nair M, Kuchipudi SV. Viruses. 2022 Jun 22;14(7):1358. | |
| Direct type I interferon signaling in hepatocytes controls malaria. Marques-da-Silva C, Peissig K, Walker MP, Shiau J, Bowers C, Kyle DE, Vijay R, Lindner SE, Kurup SP. Cell Rep. 2022 Jul 19;40(3):111098. | |
| Mechanisms underlying WNT-mediated priming of human embryonic stem cells. Yoney A, Bai L, Brivanlou AH, Siggia ED. Development. 2022 Oct 15;149(20):dev200335. | |
| Foxa2 and Pet1 Direct and Indirect Synergy Drive Serotonergic Neuronal Differentiation. Aydin B, Sierk M, Moreno-Estelles M, Tejavibulya L, Kumar N, Flames N, Mahony S, Mazzoni EO. Front Neurosci. 2022 Jun 20;16:903881. | |
| Synthetic regulatory reconstitution reveals principles of mammalian Hox cluster regulation. Pinglay S, Bulajić M, Rahe DP, Huang E, Brosh R, Mamrak NE, King BR, German S, Cadley JA, Rieber L, Easo N, Lionnet T, Mahony S, Maurano MT, Holt LJ, Mazzoni EO, Boeke JD. Science. 2022 Jul;377(6601):eabk2820. | |
| Identification and characterization of RBM12 as a novel regulator of fetal hemoglobin expression. Wakabayashi A, Kihiu M, Sharma M, Thrasher AJ, Saari MS, Quesnel-Vallières M, Abdulmalik O, Peslak SA, Khandros E, Keller CA, Giardine BM, Barash Y, Hardison RC, Shi J, Blobel GA. Blood Adv. 2022 Dec 13;6(23):5956-5968. | |
| Dual function NFI factors control fetal hemoglobin silencing in adult erythroid cells. Qin K, Huang P, Feng R, Keller CA, Peslak SA, Khandros E, Saari MS, Lan X, Mayuranathan T, Doerfler PA, Abdulmalik O, Giardine B, Chou ST, Shi J, Hardison RC, Weiss MJ, Blobel GA. Nat Genet. 2022 Jun;54(6):874-884. | |
| Lake microbiome and trophy fluctuations of the ancient hemp rettery. Iwańska O, Latoch P, Suchora M, Pidek IA, Huber M, Bubak I, Kopik N, Kovalenko M, Gąsiorowski M, Armache JP, Starosta AL. Sci Rep. 2022 May 25;12(1):8846. | |
| Standard Selection Treatments with Sulfadiazine Limit Plasmodium yoelii Host-to-Vector Transmission. Rios KT, Dickson TM, Lindner SE. mSphere. 2022 Jun 29;7(3):e0010622. | |
| Preclinical characterization and target validation of the antimalarial pantothenamide MMV693183. de Vries LE, Jansen PAM, Barcelo C, Munro J, Verhoef JMJ, Pasaje CFA, Rubiano K, Striepen J, Abla N, Berning L, Bolscher JM, Demarta-Gatsi C, Henderson RWM, Huijs T, Koolen KMJ, Tumwebaze PK, Yeo T, Aguiar ACC, Angulo-Barturen I, Churchyard A, Baum J, Fernández BC, Fuchs A, Gamo FJ, Guido RVC, Jim√©nez-Diaz MB, Pereira DB, Rochford R, Roesch C, Sanz LM, Trevitt G, Witkowski B, Wittlin S, Cooper RA, Rosenthal PJ, Sauerwein RW, Schalkwijk J, Hermkens PHH, Bonnert RV, Campo B, Fidock DA, Llinás M, Niles JC, Kooij TWA, Dechering KJ. Nat Commun. 2022 Apr 20;13(1):2158. | |
| PEGR: a flexible management platform for reproducible epigenomic and genomic research. Shao D, Kellogg GD, Nematbakhsh A, Kuntala PK, Mahony S, Pugh BF, Lai WKM. Genome Biol. 2022 Apr 19;23(1):99. | |
| Promoter competition in globin gene control. Hardison RC. Blood. 2022 Apr 7;139(14):2089-2091. | |
| The BTB transcription factors ZBTB11 and ZFP131 maintain pluripotency by repressing pro-differentiation genes. Garipler G, Lu C, Morrissey A, Lopez-Zepeda LS, Pei Y, Vidal SE, Zen Petisco Fiore AP, Aydin B, Stadtfeld M, Ohler U, Mahony S, Sanjana NE, Mazzoni EO. Cell Rep. 2022 Mar 15;38(11):110524. | |
| Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state. Nodelman IM, Das S, Faustino AM, Fried SD, Bowman GD, Armache JP. Nat Struct Mol Biol. 2022 Feb;29(2):121-129. | |
| Chemically Induced Chromosomal Interaction (CICI) method to study chromosome dynamics and its biological roles. Du M, Zou F, Li Y, Yan Y, Bai L. Nat Commun. 2022 Feb 9;13(1):757. | |
| The transcriptional regulator HDP1 controls expansion of the inner membrane complex during early sexual differentiation of malaria parasites. Campelo Morillo RA, Tong X, Xie W, Abel S, Orchard LM, Daher W, Patel DJ, Llinás M, Le Roch KG, Kafsack BFC. Nat Microbiol. 2022 Feb;7(2):289-299. | |
| Domain-adaptive neural networks improve cross-species prediction of transcription factor binding. Cochran K, Srivastava D, Shrikumar A, Balsubramani A, Hardison RC, Kundaje A, Mahony S. Genome Res. 2022 Mar;32(3):512-523. | |
| Genomic landscape of TCRαβ and TCRγδ T-large granular lymphocyte leukemia. Cheon H, Xing JC, Moosic KB, Ung J, Chan VW, Chung DS, Toro MF, Elghawy O, Wang JS, Hamele CE, Hardison RC, Olson TL, Tan SF, Feith DJ, Ratan A, Loughran TP. Blood. 2022 May 19;139(20):3058-3072. |
2021
| Heme-Edge Residues Modulate Signal Transduction within a Bifunctional Homo-Dimeric Sensor Protein. Patterson DC, Liu Y, Das S, Yennawar NH, Armache JP, Kincaid JR, Weinert EE. Biochemistry. 2021 Dec 14;60(49):3801-3812. | |
| The Plasmodium NOT1-G paralogue is an essential regulator of sexual stage maturation and parasite transmission. Hart KJ, Power BJ, Rios KT, Sebastian A, Lindner SE. PLoS Biol. 2021 Oct 21;19(10):e3001434. | |
| Probing multiple enzymatic methylation events in real time with NMR spectroscopy. Usher ET, Namitz KEW, Cosgrove MS, Showalter SA. Biophys J. 2021 Nov 2;120(21):4710-4721. | |
| Chemogenomic Fingerprints Associated with Stage-Specific Gametocytocidal Compound Action against Human Malaria Parasites. Niemand J, van Biljon R, van der Watt M, van Heerden A, Reader J, van Wyk R, Orchard L, Chibale K, Llinás M, Birkholtz LM. ACS Infect Dis. 2021 Oct 8;7(10):2904-2916. | |
| Transient Electrostatic Interactions between Fcp1 and Rap74 Bias the Conformational Ensemble of the Complex with Minimal Impact on Binding Affinity. Prieto VA, Namitz KEW, Showalter SA. J Phys Chem B. 2021 Oct 7;125(39):10917-10927. | |
| Strategies to improve pharmacogenomic-guided treatment options for patients with β-hemoglobinopathies. Patrinos GP, Chui DHK, Hardison RC, Steinberg MH. Expert Rev Hematol. 2021 Oct;14(10):883-885. | |
| Differential ligand-selective control of opposing enzymatic activities within a bifunctional c-di-GMP enzyme. Patterson DC, Ruiz MP, Yoon H, Walker JA, Armache JP, Yennawar NH, Weinert EE. Proc Natl Acad Sci U S A. 2021 Sep 7;118(36):e2100657118. | |
| CTCF and transcription influence chromatin structure re-configuration after mitosis. Zhang H, Lam J, Zhang D, Lan Y, Vermunt MW, Keller CA, Giardine B, Hardison RC, Blobel GA. Nat Commun. 2021 Aug 27;12(1):5157. | |
| DEF1: Much more than an RNA polymerase degradation factor. Akinniyi OT, Reese JC. DNA Repair (Amst). 2021 Nov;107:103202. | |
| The Penn State Protein Ladder system for inexpensive protein molecular weight markers. Santilli RT, Williamson JE 3rd, Shibata Y, Sowers RP, Fleischman AN, Tan S. Sci Rep. 2021 Aug 18;11(1):16703. | |
| A heat-shock response regulated by the PfAP2-HS transcription factor protects human malaria parasites from febrile temperatures. Tint√≥-Font E, Michel-Tod√≥ L, Russell TJ, Casas-Vila N, Conway DJ, Bozdech Z, Llinás M, Cort√©s A. Nat Microbiol. 2021 Sep;6(9):1163-1174. | |
| Physical compatibility of Normosol-R with critical care medications used in patients with COVID-19 during simulated Y-site administration. Holyk AA, Lindner AH, Lindner SE, Shippert BW. Am J Health Syst Pharm. 2022 Jan 1;79(1):e27-e33. | |
| Chemogenomics identifies acetyl-coenzyme A synthetase as a target for malaria treatment and prevention. Summers RL, Pasaje CFA, Pisco JP, Striepen J, Luth MR, Kumpornsin K, Carpenter EF, Munro JT, Lin D, Plater A, Punekar AS, Shepherd AM, Shepherd SM, Vanaerschot M, Murithi JM, Rubiano K, Akidil A, Ottilie S, Mittal N, Dilmore AH, Won M, Mandt REK, McGowen K, Owen E, Walpole C, Llinás M, Lee MCS, Winzeler EA, Fidock DA, Gilbert IH, Wirth DF, Niles JC, Baraga√±a B, Lukens AK. Cell Chem Biol. 2022 Feb 17;29(2):191-201.e8. | |
| Principles of nucleosome recognition by chromatin factors and enzymes. McGinty RK, Tan S. Curr Opin Struct Biol. 2021 Dec;71:16-26. | |
| Re-Envisioning Anti-Apicomplexan Parasite Drug Discovery Approaches. Rangel GW, Llinás M. Front Cell Infect Microbiol. 2021 Jun 11;11:691121. | |
| The Kringle of Life. Llinás M. Protein J. 2021 Aug;40(4):454-456. | |
| Single-nucleotide-level mapping of DNA regulatory elements that control fetal hemoglobin expression. Cheng L, Li Y, Qi Q, Xu P, Feng R, Palmer L, Chen J, Wu R, Yee T, Zhang J, Yao Y, Sharma A, Hardison RC, Weiss MJ, Cheng Y. Nat Genet. 2021 Jun;53(6):869-880. | |
| Intrinsically disordered substrates dictate SPOP subnuclear localization and ubiquitination activity. Usher ET, Sabri N, Rohac R, Boal AK, Mittag T, Showalter SA. J Biol Chem. 2021 Jan-Jun;296:100693. | |
| The PfAP2-G2 transcription factor is a critical regulator of gametocyte maturation. Singh S, Santos JM, Orchard LM, Yamada N, van Biljon R, Painter HJ, Mahony S, Llinás M. Mol Microbiol. 2021 May;115(5):1005-1024. | |
| Assessing relationships between chromatin interactions and regulatory genomic activities using the self-organizing map. Kunz T, Rieber L, Mahony S. Methods. 2021 May;189:12-21. | |
| Sharing biological data: why, when, and how. Wilson SL, Way GP, Bittremieux W, Armache JP, Haendel MA, Hoffman MM. FEBS Lett. 2021 Apr;595(7):847-863. | |
| H3K36 methylation reprograms gene expression to drive early gametocyte development in Plasmodium falciparum. Connacher J, Josling GA, Orchard LM, Reader J, Llinás M, Birkholtz LM. Epigenetics Chromatin. 2021 Apr 1;14(1):19. | |
| Effects of sheared chromatin length on ChIP-seq quality and sensitivity. Keller CA, Wixom AQ, Heuston EF, Giardine B, Hsiung CC, Long MR, Miller A, Anderson SM, Cockburn A, Blobel GA, Bodine DM, Hardison RC. G3 (Bethesda). 2021 Jun 17;11(6):jkab101. | |
| Frequent somatic TET2 mutations in chronic NK-LGL leukemia with distinct patterns of cytopenias. Olson TL, Cheon H, Xing JC, Olson KC, Paila U, Hamele CE, Neelamraju Y, Shemo BC, Schmachtenberg M, Sundararaman SK, Toro MF, Keller CA, Farber EA, Onengut-Gumuscu S, Garrett-Bakelman FE, Hardison RC, Feith DJ, Ratan A, Loughran TP. Blood. 2021 Aug 26;138(8):662-673. | |
| PfMFR3: A Multidrug-Resistant Modulator in Plasmodium falciparum. Rocamora F, Gupta P, Istvan ES, Luth MR, Carpenter EF, K√ºmpornsin K, Sasaki E, Calla J, Mittal N, Carolino K, Owen E, Llinás M, Ottilie S, Goldberg DE, Lee MCS, Winzeler EA. ACS Infect Dis. 2021 Apr 9;7(4):811-825. | |
| A high-resolution protein architecture of the budding yeast genome. Rossi MJ, Kuntala PK, Lai WKM, Yamada N, Badjatia N, Mittal C, Kuzu G, Bocklund K, Farrell NP, Blanda TR, Mairose JD, Basting AV, Mistretta KS, Rocco DJ, Perkinson ES, Kellogg GD, Mahony S, Pugh BF. Nature. 2021 Apr;592(7853):309-314. | |
| S3V2-IDEAS: a package for normalizing, denoising and integrating epigenomic datasets across different cell types. Xiang G, Giardine BM, Mahony S, Zhang Y, Hardison RC. Bioinformatics. 2021 Sep 29;37(18):3011-3013. | |
| MalDA, Accelerating Malaria Drug Discovery. Yang T, Ottilie S, Istvan ES, Godinez-Macias KP, Lukens AK, Baraga√±a B, Campo B, Walpole C, Niles JC, Chibale K, Dechering KJ, Llinás M, Lee MCS, Kato N, Wyllie S, McNamara CW, Gamo FJ, Burrows J, Fidock DA, Goldberg DE, Gilbert IH, Wirth DF, Winzeler EA; Malaria Drug Accelerator Consortium. Trends Parasitol. 2021 Jun;37(6):493-507. | |
| Regulation of Promoter Proximal Pausing of RNA Polymerase II in Metazoans. Dollinger R, Gilmour DS. J Mol Biol. 2021 Jul 9;433(14):166897. | |
| Limited window for donation of convalescent plasma with high live-virus neutralizing antibody titers for COVID-19 immunotherapy. Gontu A, Srinivasan S, Salazar E, Nair MS, Nissly RH, Greenawalt D, Bird IM, Herzog CM, Ferrari MJ, Poojary I, Katani R, Lindner SE, Minns AM, Rossi R, Christensen PA, Castillo B, Chen J, Eagar TN, Yi X, Zhao P, Leveque C, Olsen RJ, Bernard DW, Gollihar J, Kuchipudi SV, Musser JM, Kapur V. Commun Biol. 2021 Feb 24;4(1):267. | |
| Distinct properties and functions of CTCF revealed by a rapidly inducible degron system. Luan J, Xiang G, Gómez-García PA, Tome JM, Zhang Z, Vermunt MW, Zhang H, Huang A, Keller CA, Giardine BM, Zhang Y, Lan Y, Lis JT, Lakadamyali M, Hardison RC, Blobel GA. Cell Rep. 2021 Feb 23;34(8):108783. | |
| Dynamic CTCF binding directly mediates interactions among cis-regulatory elements essential for hematopoiesis. Qi Q, Cheng L, Tang X, He Y, Li Y, Yee T, Shrestha D, Feng R, Xu P, Zhou X, Pruett-Miller S, Hardison RC, Weiss MJ, Cheng Y. Blood. 2021 Mar 11;137(10):1327-1339. | |
| Atypical Molecular Basis for Drug Resistance to Mitochondrial Function Inhibitors in Plasmodium falciparum. Painter HJ, Morrisey JM, Mather MW, Orchard LM, Luck C, Smilkstein MJ, Riscoe MK, Vaidya AB, Llinás M. Antimicrob Agents Chemother. 2021 Feb 17;65(3):e02143-20. | |
| The Ada2/Ada3/Gcn5/Sgf29 histone acetyltransferase module. Espinola-Lopez JM, Tan S. Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194629. | |
| Artemisinin-resistant K13 mutations rewire Plasmodium falciparum's intra-erythrocytic metabolic program to enhance survival. Mok S, Stokes BH, Gn√§dig NF, Ross LS, Yeo T, Amaratunga C, Allman E, Solyakov L, Bottrill AR, Tripathi J, Fairhurst RM, Llinás M, Bozdech Z, Tobin AB, Fidock DA. Nat Commun. 2021 Jan 22;12(1):530. | |
| Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation. Valencia-Sánchez MI, De Ioannes P, Wang M, Truong DM, Lee R, Armache JP, Boeke JD, Armache KJ. Science. 2021 Jan 22;371(6527):eabc6663. | |
| ZNF410 Uniquely Activates the NuRD Component CHD4 to Silence Fetal Hemoglobin Expression. Lan X, Ren R, Feng R, Ly LC, Lan Y, Zhang Z, Aboreden N, Qin K, Horton JR, Grevet JD, Mayuranathan T, Abdulmalik O, Keller CA, Giardine B, Hardison RC, Crossley M, Weiss MJ, Cheng X, Shi J, Blobel GA. Mol Cell. 2021 Jan 21;81(2):239-254.e8. | |
| Multistage and transmission-blocking targeted antimalarials discovered from the open-source MMV Pandemic Response Box. Reader J, van der Watt ME, Taylor D, Le Manach C, Mittal N, Ottilie S, Theron A, Moyo P, Erlank E, Nardini L, Venter N, Lauterbach S, Bezuidenhout B, Horatscheck A, van Heerden A, Spillman NJ, Cowell AN, Connacher J, Opperman D, Orchard LM, Llinás M, Istvan ES, Goldberg DE, Boyle GA, Calvo D, Mancama D, Coetzer TL, Winzeler EA, Duffy J, Koekemoer LL, Basarab G, Chibale K, Birkholtz LM. Nat Commun. 2021 Jan 11;12(1):269. | |
| Thermodynamic modeling of genome-wide nucleosome depleted regions in yeast. Kharerin H, Bai L. PLoS Comput Biol. 2021 Jan 11;17(1):e1008560. | |
| Clinically relevant updates of the HbVar database of human hemoglobin variants and thalassemia mutations. Giardine BM, Joly P, Pissard S, Wajcman H, K Chui DH, Hardison RC, Patrinos GP. Nucleic Acids Res. 2021 Jan 8;49(D1):D1192-D1196. | |
| An interpretable bimodal neural network characterizes the sequence and preexisting chromatin predictors of induced transcription factor binding. Srivastava D, Aydin B, Mazzoni EO, Mahony S. Genome Biol. 2021 Jan 7;22(1):20. |
2020
| Ash1 and Tup1 dependent repression of the Saccharomyces cerevisiae HO promoter requires activator-dependent nucleosome eviction. Parnell EJ, Parnell TJ, Yan C, Bai L, Stillman DJ. PLoS Genet. 2020 Dec 31;16(12):e1009133. |
| Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch. Lehmann LC, Bacic L, Hewitt G, Brackmann K, Sabantsev A, Gaullier G, Pytharopoulou S, Degliesposti G, Okkenhaug H, Tan S, Costa A, Skehel JM, Boulton SJ, Deindl S. Cell Rep. 2020 Dec 22;33(12):108529. |
| Mapping invisible epitopes by NMR spectroscopy. Usher ET, Showalter SA. J Biol Chem. 2020 Dec 18;295(51):17411-17412. |
| An adjunctive therapy administered with an antibiotic prevents enrichment of antibiotic-resistant clones of a colonizing opportunistic pathogen. Morley VJ, Kinnear CL, Sim DG, Olson SN, Jackson LM, Hansen E, Usher GA, Showalter SA, Pai MP, Woods RJ, Read AF. Elife. 2020 Dec 1;9:e58147. |
| A map of cis-regulatory elements and 3D genome structures in zebrafish. Yang H, Luan Y, Liu T, Lee HJ, Fang L, Wang Y, Wang X, Zhang B, Jin Q, Ang KC, Xing X, Wang J, Xu J, Song F, Sriranga I, Khunsriraksakul C, Salameh T, Li D, Choudhary MNK, Topczewski J, Wang K, Gerhard GS, Hardison RC, Wang T, Cheng KC, Yue F. Nature. 2020 Dec;588(7837):337-343. |
| Elucidating the Role of Microprocessor Protein DGCR8 in Bending RNA Structures. Pabit SA, Chen YL, Usher ET, Cook EC, Pollack L, Showalter SA. Biophys J. 2020 Dec 15;119(12):2524-2536. |
| Increased circulation time of Plasmodium falciparum underlies persistent asymptomatic infection in the dry season. Andrade CM, Fleckenstein H, Thomson-Luque R, Doumbo S, Lima NF, Anderson C, Hibbert J, Hopp CS, Tran TM, Li S, Niangaly M, Cisse H, Doumtabe D, Skinner J, Sturdevant D, Ricklefs S, Virtaneva K, Asghar M, Homann MV, Turner L, Martins J, Allman EL, N'Dri ME, Winkler V, Llinás M, Lavazec C, Martens C, F√§rnert A, Kayentao K, Ongoiba A, Lavstsen T, Os√≥rio NS, Otto TD, Recker M, Traore B, Crompton PD, Portugal S. Nat Med. 2020 Dec;26(12):1929-1940. |
| Differential abilities to engage inaccessible chromatin diversify vertebrate Hox binding patterns. Bulajić M, Srivastava D, Dasen JS, Wichterle H, Mahony S, Mazzoni EO. Development. 2020 Nov 23;147(22):dev194761. |
| Definition of constitutive and stage-enriched promoters in the rodent malaria parasite, Plasmodium yoelii. Bowman LM, Finger LE, Hart KJ, Lindner SE. Malar J. 2020 Nov 23;19(1):424. |
| Alignment and quantification of ChIP-exo crosslinking patterns reveal the spatial organization of protein-DNA complexes. Yamada N, Rossi MJ, Farrell N, Pugh BF, Mahony S. Nucleic Acids Res. 2020 Nov 18;48(20):11215-11226. |
| Alteration of genome folding via contact domain boundary insertion. Zhang D, Huang P, Sharma M, Keller CA, Giardine B, Zhang H, Gilgenast TG, Phillips-Cremins JE, Hardison RC, Blobel GA. Nat Genet. 2020 Oct;52(10):1076-1087. Determination of human identity from Anopheles stephensi mosquito blood meals using direct amplification and massively parallel sequencing. Gray SL, Tiedge TM, Butkus JM, Earp TJ, Lindner SE, Roy R. Forensic Sci Int Genet. 2020 Sep;48:102347. |
| In silico identification of Drosophila melanogaster genes encoding RNA polymerase subunits. Marygold SJ, Alic N, Gilmour DS, Grewal SS. MicroPubl Biol. 2020 Oct 20;2020:10.17912/micropub.biology.000320. |
| HRI depletion cooperates with pharmacologic inducers to elevate fetal hemoglobin and reduce sickle cell formation. Peslak SA, Khandros E, Huang P, Lan X, Geronimo CL, Grevet JD, Abdulmalik O, Zhang Z, Giardine BM, Keller CA, Shi J, Hardison RC, Blobel GA. Blood Adv. 2020 Sep 22;4(18):4560-4572. |
| The natural function of the malaria parasite's chloroquine resistance transporter. Shafik SH, Cobbold SA, Barkat K, Richards SN, Lancaster NS, Llinás M, Hogg SJ, Summers RL, McConville MJ, Martin RE. Nat Commun. 2020 Aug 6;11(1):3922. |
| Expanded encyclopaedias of DNA elements in the human and mouse genomes. ENCODE Project Consortium; Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, Kaul R, Halow J, Van Nostrand EL, Freese P, Gorkin DU, Shen Y, He Y, Mackiewicz M, Pauli-Behn F, Williams BA, Mortazavi A, Keller CA, Zhang XO, Elhajjajy SI, Huey J, Dickel DE, Snetkova V, Wei X, Wang X, Rivera-Mulia JC, Rozowsky J, Zhang J, Chhetri SB, Zhang J, Victorsen A, White KP, Visel A, Yeo GW, Burge CB, Lécuyer E, Gilbert DM, Dekker J, Rinn J, Mendenhall EM, Ecker JR, Kellis M, Klein RJ, Noble WS, Kundaje A, Guigó R, Farnham PJ, Cherry JM, Myers RM, Ren B, Graveley BR, Gerstein MB, Pennacchio LA, Snyder MP, Bernstein BE, Wold B, Hardison RC, Gingeras TR, Stamatoyannopoulos JA, Weng Z. Nature. 2020 Jul;583(7818):699-710. |
| A Cambrian origin for globin gene regulation. Hardison RC. Blood. 2020 Jul 16;136(3):261-262. |
| PEGR: a management platform for ChIP-based next generation sequencing pipelines. Shao D, Kellogg G, Mahony S, Lai W, Pugh BF. PEARC20 (2020). 2020 Jul;2020:285-292. |
| The changing mouse embryo transcriptome at whole tissue and single-cell resolution. He P, Williams BA, Trout D, Marinov GK, Amrhein H, Berghella L, Goh ST, Plajzer-Frick I, Afzal V, Pennacchio LA, Dickel DE, Visel A, Ren B, Hardison RC, Zhang Y, Wold BJ. Nature. 2020 Jul;583(7818):760-767. |
| Perspectives on ENCODE. ENCODE Project Consortium; Snyder MP, Gingeras TR, Moore JE, Weng Z, Gerstein MB, Ren B, Hardison RC, Stamatoyannopoulos JA, Graveley BR, Feingold EA, Pazin MJ, Pagan M, Gilchrist DA, Hitz BC, Cherry JM, Bernstein BE, Mendenhall EM, Zerbino DR, Frankish A, Flicek P, Myers RM. Nature. 2020 Jul;583(7818):693-698. |
| Inhibition of Resistance-Refractory P. falciparum Kinase PKG Delivers Prophylactic, Blood Stage, and Transmission-Blocking Antiplasmodial Activity. Vanaerschot M, Murithi JM, Pasaje CFA, Ghidelli-Disse S, Dwomoh L, Bird M, Spottiswoode N, Mittal N, Arendse LB, Owen ES, Wicht KJ, Siciliano G, B√∂sche M, Yeo T, Kumar TRS, Mok S, Carpenter EF, Giddins MJ, Sanz O, Ottilie S, Alano P, Chibale K, Llinás M, Uhlemann AC, Delves M, Tobin AB, Doerig C, Winzeler EA, Lee MCS, Niles JC, Fidock DA. Cell Chem Biol. 2020 Jul 16;27(7):806-816.e8. |
| Refining the transcriptome of the human malaria parasite Plasmodium falciparum using amplification-free RNA-seq. Chappell L, Ross P, Orchard L, Russell TJ, Otto TD, Berriman M, Rayner JC, Llinás M. BMC Genomics. 2020 Jun 8;21(1):395. |
| Crystal Structure of the LSD1/CoREST Histone Demethylase Bound to Its Nucleosome Substrate. Kim SA, Zhu J, Yennawar N, Eek P, Tan S. Mol Cell. 2020 Jun 4;78(5):903-914.e4. |
| A Role for Mediator Core in Limiting Coactivator Recruitment in Saccharomyces cerevisiae. Yarrington RM, Yu Y, Yan C, Bai L, Stillman DJ. Genetics. 2020 Jun;215(2):407-420. |
| The HRI-regulated transcription factor ATF4 activates BCL11A transcription to silence fetal hemoglobin expression. Huang P, Peslak SA, Lan X, Khandros E, Yano JA, Sharma M, Keller CA, Giardine B, Qin K, Abdulmalik O, Hardison RC, Shi J, Blobel GA. Blood. 2020 Jun 11;135(24):2121-2132. |
| Sequence and chromatin determinants of transcription factor binding and the establishment of cell type-specific binding patterns. Srivastava D, Mahony S. Biochim Biophys Acta Gene Regul Mech. 2020 Jun;1863(6):194443. |
| Genetic ablation of the mitoribosome in the malaria parasite Plasmodium falciparum sensitizes it to antimalarials that target mitochondrial functions. Ling L, Mulaka M, Munro J, Dass S, Mather MW, Riscoe MK, Llinás M, Zhou J, Ke H. J Biol Chem. 2020 May 22;295(21):7235-7248. |
| Understanding heterogeneity of fetal hemoglobin induction through comparative analysis of F and A erythroblasts. Khandros E, Huang P, Peslak SA, Sharma M, Abdulmalik O, Giardine BM, Zhang Z, Keller CA, Hardison RC, Blobel GA. Blood. 2020 May 28;135(22):1957-1968. |
| Reviving Protein-Observed 19F Lineshape Analysis for Deep Insight into Protein-Ligand Binding Events. Showalter SA. Biophys J. 2020 May 19;118(10):2333-2335. |
| S3norm: simultaneous normalization of sequencing depth and signal-to-noise ratio in epigenomic data. Xiang G, Keller CA, Giardine B, An L, Li Q, Zhang Y, Hardison RC. Nucleic Acids Res. 2020 May 7;48(8):e43. |
| Ultrasound-Guided Cytosolic Protein Delivery via Transient Fluorous Masks. Sloand JN, Nguyen TT, Zinck SA, Cook EC, Zimudzi TJ, Showalter SA, Glick AB, Simon JC, Medina SH. ACS Nano. 2020 Apr 28;14(4):4061-4073. |
| An integrative view of the regulatory and transcriptional landscapes in mouse hematopoiesis. Xiang G, Keller CA, Heuston E, Giardine BM, An L, Wixom AQ, Miller A, Cockburn A, Sauria MEG, Weaver K, Lichtenberg J, Göttgens B, Li Q, Bodine D, Mahony S, Taylor J, Blobel GA, Weiss MJ, Cheng Y, Yue F, Hughes J, Higgs DR, Zhang Y, Hardison RC. Genome Res. 2020 Mar;30(3):472-484. |
| Dissecting the role of PfAP2-G in malaria gametocytogenesis. Josling GA, Russell TJ, Venezia J, Orchard L, van Biljon R, Painter HJ, Llinás M. Nat Commun. 2020 Mar 20;11(1):1503. |
| ChExMix: A Method for Identifying and Classifying Protein-DNA Interaction Subtypes. Yamada N, Kuntala PK, Pugh BF, Mahony S. J Comput Biol. 2020 Mar;27(3):429-435. |
| A Novel Antiparasitic Compound Kills Ring-Stage Plasmodium falciparum and Retains Activity Against Artemisinin-Resistant Parasites. Clements RL, Streva V, Dumoulin P, Huang W, Owens E, Raj DK, Burleigh B, Llinás M, Winzeler EA, Zhang Q, Dvorin JD. J Infect Dis. 2020 Mar 2;221(6):956-962. |
| Combining Stage Specificity and Metabolomic Profiling to Advance Antimalarial Drug Discovery. Murithi JM, Owen ES, Istvan ES, Lee MCS, Ottilie S, Chibale K, Goldberg DE, Winzeler EA, Llinás M, Fidock DA, Vanaerschot M. Cell Chem Biol. 2020 Feb 20;27(2):158-171.e3. |
| A single-nucleotide polymorphism in a Plasmodium berghei ApiAP2 transcription factor alters the development of host immunity. Akkaya M, Bansal A, Sheehan PW, Pena M, Molina-Cruz A, Orchard LM, Cimperman CK, Qi CF, Ross P, Yazew T, Sturdevant D, Anzick SL, Thiruvengadam G, Otto TD, Billker O, Llinás M, Miller LH, Pierce SK. Sci Adv. 2020 Feb 5;6(6):eaaw6957. |
| Systematic integration of GATA transcription factors and epigenomes via IDEAS paints the regulatory landscape of hematopoietic cells. Hardison RC, Zhang Y, Keller CA, Xiang G, Heuston EF, An L, Lichtenberg J, Giardine BM, Bodine D, Mahony S, Li Q, Yue F, Weiss MJ, Blobel GA, Taylor J, Hughes J, Higgs DR, Göttgens B. IUBMB Life. 2020 Jan;72(1):27-38. |
2019
| Chromatin structure dynamics during the mitosis-to-G1 phase transition Haoyue Zhang, Daniel J. Emerson, Thomas G. Gilgenast, Katelyn R. Titus, Yemin Lan, Peng Huang, Di Zhang, Hongxin Wang, Cheryl A. Keller, Belinda Giardine, Ross C. Hardison, Jennifer E. Phillips-Cremins, Gerd A. Blobel Nature, 576, 158–162, 2019 |  |
| Sequence and chromatin determinants of transcription factor binding and the establishment of cell type-specific binding patterns Divyanshi Srivastava, Shaun Mahony Elsevier, 19:194443, 2019 |  |
| Systematic integration of GATA transcription factors andepigenomes via IDEAS paints the regulatory landscape ofhematopoietic cells Ross C. Hardison, Yu Zhang, Cheryl A. Keller, Guanjue Xiang, Elisabeth F. Heuston, Lin An, Jens Lichtenberg, Belinda M. Giardine, David Bodine, Shaun Mahony, Qunhua Li, Feng Yue, Mitchell J. Weiss, Gerd A. Blobel, James Taylor, Jim Hughes, Douglas R. Higgs, Berthold Göttgens IUBMB Life, 72:27–38, 2019 |  |
| Enhancement of LacI binding in vivo Fan Zou, Manyu Du, Hengye Chen, Lu Bai Molecular and Cellular Biology, 39:e00146-19., 2019 |  |
| Existence, Transition, and Propagation of Intermediate Silencing States in Ribosomal DNA Manyu Du, Seth Kodner, Lu Bai Nucleic Acids Research, 9609–9618, 2019 |  |
| Direct prediction of regulatory elements from partial data without imputation Yu Zhang, Shaun Mahony PLOS Computational Biology, 15:1007399, 2019 |  |
| Joint inference and alignment of genome structures enables characterization of compartment-independent reorganization across cell types Lila Rieber, Shaun Mahony Epigenetics & Chromatin, 139-140, 2019 |  |
| Single-particle cryo-EM: beyond the resolution Jean-Paul Armache, Yifan Cheng Biology & Biochemistry, 864-866, 2019 |  |
| Cryo-EM of multiple cage architectures reveals a universal mode of clathrin self-assembly Kyle L. Morris, Joseph R. Jones, Mary Halebian, Shenping Wu, Michael Baker, Jean-Paul Armache, Amaurys Avila Ibarra, Richard B. Sessions, Alexander D. Cameron, Yifan Cheng, Corinne J. Smith Nature Structural & Molecular Biology, 890-898, 2019 |  |
| Single-molecule FRET method to investigate the dynamics of transcriptionelongation through the nucleosome by RNA polymerase II Jaehyoun Lee, J. Brooks Crickard, Joseph C. Reese, Tae-Hee Lee Elsevier, 159-160: 51-58, 2019 |  |
| Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination Marco Igor Valencia-Sánchez, Pablo De Ioannes, Miao Wang, Nikita Vasilyev, Ruoyu Chen, Evgeny Nudler, Jean-Paul Armache, and Karim-Jean Armache Molecular Cell, 74:1010-1019, 2019 |  |
| Cryo-EM structures of remodeler- nucleosome intermediates suggest allosteric control through the nucleosome Jean Paul Armache, Nathan Gamarra, Stephanie L Johnson, John D Leonard, Shenping Wu, Geeta J Narlikar and Yifan Cheng eLife, 8:e46057, 2019 |  |
| Dissociation rate compensation mechanism for budding yeast pioneer transcription factors Benjamin T Donovan, Hengye Chen, Caroline Jipa, Lu Bai and Michael G Poirier eLife, 8:e46057, 2019 |  |
| Using time-lapse fluorescence microscopy to study gene regulation Fan Zou and Lu Bai Methods, 159-160:138-145, 2019 |  |
| Genetic analysis of the RNA polymerase II CTD in Drosophila Feiyue Lu and David S. Gilmour Methods, 159-160:129-137, 2019 |  |
| The C-Terminal Domain of RNA Polymerase II Is a Multivalent Targeting Sequence that Supports Drosophila Development with Only Consensus Heptad Feiyue Lu, Bede Portz and David S. Gilmour Molecular Cell, 73:1232-1242, 2019 |  |
| Transcriptional Burst Initiation and Polymerase Pause Release Are Key Control Points of Transcriptional Regulation Caroline R. Bartman, Nicole Hamagami, Cheryl A. Keller, Belinda Giardine, Ross C. Hardison, Gerd A. Blobel and Arjun Raj Molecular Cell, 73:519-532, 2019 |  |
| Interrogating Histone Acetylation and BRD4 as Mitotic Bookmarks of Transcription Vivek Behera, Aaron J. Stonestrom, Nicole Hamagami, Chris C. Hsiung, Cheryl A. Keller, Belinda Giardine, Simone Sidoli, Zuo-Fei Yuan, Natarajan V. Bhanu, Michael T. Werner, Hongxin Wang, Benjamin A. Garcia, Ross C. Hardison and Gerd A. Blobel Cell Reports, 27:400-415, 2019 |  |
| Proneural factors Ascl1 and Neurog2 contribute to neuronal subtype identities by establishing distinct chromatin landscapes Begüm Aydin, Akshay Kakumanu, Mary Rossillo, Mireia Moreno-Estellés, Görkem Garipler, Niels Ringstad, Nuria Flames, Shaun Mahony and Esteban O. Mazzoni Nature Neuroscience, 22:897-908, 2019 |  |
| Biochemical methods to characterize RNA polymerase II elongation T complexes J. Brooks Crickard and Joseph C. Reese Methods, 159-160:70-81, 2019 |  |
| Ccr4–Not maintains genomic integrity by controlling the ubiquitylation and degradation of arrested RNAPII Haoyang Jiang, Marley Wolgast, Laura M. Beebe and Joseph C. Reese Genes & Development, 33:705-717, 2019 |  |
| Solution Ensemble of the C‐Terminal Domain from the Transcription Factor Pdx1 Resembles an Excluded Volume Polymer Erik C. Cook, Debashish Sahu, Monique Bastidas and Scott A. Showalter J. Physical Chemistry B, 123:106-116, 2019 |  |
2018
| Systematic Study of Nucleosome-Displacing Factors in Budding Yeast Chao Yan, Hengye Chen and Lu Bai Molecular Cell, 71:294-305, 2018 |  |
| GFZF, a Glutathione S-Transferase Protein Implicated in Cell Cycle Regulation and Hybrid Inviability, Is a Transcriptional Coactivator Douglas G. Baumann, Mu-Shui Dai, Hua Lu and David S. Gilmour Mol. Cell. Biol., 38:e00476-17, 2018 |  |
| Exploiting genetic variation to uncover rules of transcription factor binding and chromatin accessibility Vivek Behera, Perry Evans, Carolyne J. Face, Nicole Hamagami, Laavanya Sankaranarayanan, Cheryl A. Keller, Belinda Giardine, Kai Tan, Ross C. Hardison, Junwei Shi & Gerd A. Blobe Nature Communications, 9:782, 2018 | 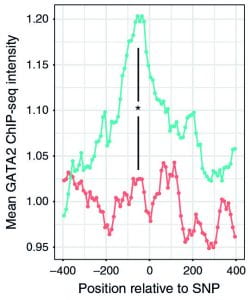 |
| Domain-focused CRISPR screen identifies HRI as a fetal hemoglobin regulator in human erythroid cells Jeremy D. Grevet, Xianjiang Lan, Nicole Hamagami, Christopher R. Edwards, Laavanya Sankaranarayanan, Xinjun Ji, Saurabh K. Bhardwaj, Carolyne J. Face, David F. Posocco, Osheiza Abdulmalik, Cheryl A. Keller, Belinda Giardine, Simone Sidoli, Ben A. Garcia, Stella T. Chou, Stephen A. Liebhaber, Ross C. Hardison, Junwei Shi and Gerd A. Blobel Science, 361:285-290, 2018 |  |
| Establishment of regulatory elements during erythro-megakaryopoiesis identifies hematopoietic lineage-commitment points Elisabeth F. Heuston, Cheryl A. Keller, Jens Lichtenberg, Belinda Giardine, Stacie M. Anderson, NIH Intramural Sequencing Center, Ross C. Hardison and David M. Bodine Epigenetics & Chromatin, 11:22, 2018 |  |
| Open-source discovery of chemical leads for next-generation chemoprotective antimalarials Yevgeniya Antonova-Koch et al Science, 362:1129, 2018 |  |
| Specific Inhibition of the Bifunctional Farnesyl/ Geranylgeranyl Diphosphate Synthase in Malaria Parasites via a New Small-Molecule Binding Site Jolyn E. Gisselberg, Zachary Herrera, Lindsey M. Orchard, Manuel Llinás and Ellen Yeh Cell Chemical Biology, 25:185-193, 2018 |  |
| Genome-wide real-time in vivo transcriptional dynamics during Plasmodium falciparum blood-stage development Heather J. Painter, Neo Christopher Chung, Aswathy Sebastian, Istvan Albert, John D. Storey, & Manuel Llinás Nature Communications, 9:2656, 2018 |  |
| Characterizing protein–DNA binding event subtypes in ChIP-exo data Naomi Yamada, William K. M. Lai, Nina Farrell, B. Franklin Pugh and Shaun Mahony Bioinformatics, 35:903-913, 2018 |  |
| Genome-wide determinants of sequence-specific DNA binding of general regulatory factors Matthew J. Rossi, William K.M. Lai and B. Franklin Pugh Genome Research, 28:497-508, 2018 |  |
| Simplified ChIP-exo assays Matthew J. Rossi, William K.M. Lai & B. Franklin Pugh Nature Communications, 9:2842, 2018 |  |
| Widespread and precise reprogramming of yeast protein–genome interactions in response to heat shock Vinesh Vinayachandran, Rohit Reja, Matthew J. Rossi, Bongsoo Park, Lila Rieber, Chitvan Mittal, Shaun Mahony and B. Franklin Pugh Genome Research, 28:357=366, 2018 |  |
| Genome-Wide Mapping of Decay Factor–mRNA Interactions in Yeast Identifies Nutrient-Responsive Transcripts as Targets of the Deadenylase Ccr4 Jason E. Miller, Liye Zhang, Haoyang Jiang, Yunfei Li, B. Franklin Pugh and Joseph C. Reese G3, 8:315-330, 2018 |  |
| The Use of 13C Direct-Detect NMR to Characterize Flexible and Disordered Proteins Erik C. Cook, Grace A. Usher and Scott A. Showalter Methods in Enzymology, 611:81-100, 2018 |  |
| Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2 Jian Sun, Marcin Paduch, Sang-Ah Kim, Ryan M. Kramer, Adam F. Barrios, Vincent Lu, Judy Luke, Svitlana Usatyuk, Anthony A. Kossiakoff and Song Tan PNAS, 115:10010-10015, 2018 |  |
2017
| Three distinct mechanisms of long-distance modulation of gene expression in yeast Manyu Du,, Qian Zhang and Lu Bai PLOS Genetics, 13:e1006736, 2017 |  |
| A sequence-specific core promoter-binding transcription factor recruits TRF2 to coordinately transcribe ribosomal protein genes Douglas G. Baumann and David S. Gilmour Nucleic Acids Research, 45:10481-10491, 2017 |  |
| Structural heterogeneity in the intrinsically disordered RNA polymerase II C-terminal domain Bede Portz, Feiyue Lu,, Eric B. Gibbs, Joshua E. Mayfield, M. Rachel Mehaffey, Yan Jessie Zhang, Jennifer S. Brodbelt, Scott A. Showalter & David S. Gilmour Nature Communications, 8:15231, 2017 |  |
| Identification of Regions in the Spt5 Subunit of DRB Sensitivity-inducing Factor (DSIF) That Are Involved in Promoter-proximal Pausing Yijun Qiu and David S. Gilmour J. Biol. Chem., 292:5555-5570, 2017 |  |
| Comparative analysis of three-dimensional chromosomal architecture identifies a novel fetal hemoglobin regulatory element Peng Huang, Cheryl A. Keller, Belinda Giardine, Jeremy D. Grevet, James O.J. Davies, Jim R. Hughes, Ryo Kurita, Yukio Nakamura, Ross C. Hardison and Gerd A. Blobel Genes & Development, 31:1704-1713, 2017 |  |
| Accurate and reproducible functional maps in 127 human cell types via 2D genome segmentation Yu Zhang and Ross C. Hardison Nucleic Acids Research, 45:9823-9836, 2017 |  |
| Capturing in vivo RNA transcriptional dynamics from the malaria parasite Plasmodium falciparum Heather J. Painter, Manuela Carrasquilla and Manuel Llinás Genome Research, 27:1074-1086, 2017 |  |
| Red Blood Cell Invasion by the Malaria Parasite Is Coordinated by the PfAP2-I Transcription Factor Joana Mendonca Santos, Gabrielle Josling, Philipp Ross, Preeti Joshi, Lindsey Orchard, Tracey Campbell, Ariel Schieler, Ileana M. Cristea, and Manuel Llinás Cell Host & Microbe, 731-741.e10, 2017 |  |
| Deconvolving sequence features that discriminate between overlapping regulatory annotations Akshay Kakumanu, Silvia Velasco, Esteban Mazzoni, Shaun Mahony PLOS Computational Biology, 13:e1005795, 2017 |  |
| miniMDS: 3D structural inference from high-resolution Hi-C data Lila Rieber and Shaun Mahony Bioinformatics, 33:i261–i266, 2017 |  |
| A Multi-step Transcriptional and Chromatin State Cascade Underlies Motor Neuron Programming from Embryonic Stem Cells Silvia Velasco, Mahmoud M. Ibrahim, Akshay Kakumanu, Görkem Garipler, Begu€m Aydin, Mohamed Ahmed Al-Sayegh, Antje Hirsekorn, Farah Abdul-Rahman, Rahul Satija, Uwe Ohler, Shaun Mahony and Esteban O. Mazzoni Cell Stem Cell, 20:205-217, 2017 |  |
| The elongation factor Spt4/5 regulates RNA polymerase II transcription through the nucleosome John B. Crickard, Jaehyoun Lee, Tae-Hee Lee and Joseph C. Reese Nucleic Acids Research, 45:6362-6374, 2017 |  |
| Phosphorylation induces sequence-specific conformational switches in the RNA polymerase II C-terminal domain Eric B. Gibbs, Feiyue Lu, Bede Portz, Michael J. Fisher, Brenda P. Medellin, Tatiana N. Laremore, Yan Jessie Zhang, David S. Gilmour & Scott A. Showalter Nature Communications, 8:15233, 2017 |  |
| Substrate Specificity of the Kinase P-TEFb towards the RNA Polymerase II C-Terminal Domain Eric B. Gibbs, Tatiana N. Laremore, Grace A. Usher, Bede Portz, Erik C. Cook and Scott A. Showalter Biophysical Journal, 113:1909-1911, 2017 |  |
| Engineering double-stranded RNA binding activity into the Drosha double-stranded RNA binding domain results in a loss of microRNA processing function Joshua C. Kranick, Durga M. Chadalavada, Debashish Sahu and Scott A. Showalter PLOS One, 12:e0182445, 2017 |  |
| The pPSU Plasmids for Generating DNA Molecular Weight Markers Ryan C. Henrici, Turner J. Pecen, James L. Johnston & Song Tan Scientific Reports, 7:2438, 2017 |  |
2016
| Regulation of cell-to-cell variability in divergent gene expression Chao Yan,, Shuyang Wu, Christopher Pocetti & Lu Bai Nature Communications, 7:11099, 2016 |  |
| Interallelic interaction and gene regulation in budding yeast Daoyong Zhang and Lu Bai PNAS, 113:4428-4433, 2016 |  |
| A hyperactive transcriptional state marks genome reactivation at the mitosis–G1 transition Chris C.-S. Hsiung, Caroline R. Bartman, Peng Huang, Paul Ginart, Aaron J. Stonestrom, Cheryl A. Keller, Carolyne Face, Kristen S. Jahn, Perry Evans, Laavanya Sankaranarayanan, Belinda Giardine, Ross C. Hardison, Arjun Raj, and Gerd A. Blobel Genes & Development, 30:1423-14439, 2016 |  |
| Unlinking an lncRNA from Its Associated cis Element Vikram R. Paralkar, Cristian C. Taborda, Peng Huang, Yu Yao, Andrew V. Kossenkov, Rishi Prasad, Jing Luan, James O.J. Davies, Jim R. Hughes, Ross C. Hardison, Gerd A. Blobel, and Mitchell J. Weiss Molecular Cell, 62:104-110, 2016 |  |
| Jointly characterizing epigenetic dynamics across multiple human cell types Yu Zhang, Lin An, Feng Yue and Ross C. Hardison Nucleic Acids Research, 44:6721-6731, 2016 | 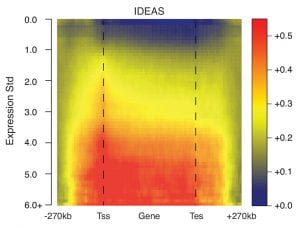 |
| Proteome-wide analysis reveals widespread lysine acetylation of major protein complexes in the malaria parasite Simon A. Cobbold, Joana M. Santos, Alejandro Ochoa, David H. Perlman & Manuel Llinás Scientific Reports, 6:19722, 2016 |  |
| The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation Makiko Iwafuchi-Doi, Greg Donahue, Akshay Kakumanu, Jason A. Watts, Shaun Mahony, B. Franklin Pugh, Dolim Lee, Klaus H. Kaestner and Kenneth S. Zaret Molecular Cell, 62:79-91, 2016 |  |
| Genome-Wide Organization of GATA1 and TAL1 Determined at High Resolution G. Celine Han, Vinesh Vinayachandran, Alain R. Bataille, Bongsoo Park, Ka Yim Chan-Salis, Cheryl A. Keller, Maria Long, Shaun Mahony, Ross C. Hardison, B. Franklin Pugh Mol Cell Biol, 36:157-172, 2016 |  |
| Genomic Nucleosome Organization Reconstituted with Pure Proteins Nils Krietenstein, Megha Wal, Shinya Watanabe, Bongsoo Park, Craig L. Peterson, B. Franklin Pugh, and Philipp Korber Cell, 167:709-721, 2016 |  |
| Biochemical Analysis of Yeast Suppressor of Ty 4/5 (Spt4/5) Reveals the Importance of Nucleic Acid Interactions in the Prevention of RNA Polymerase II Arrest J. Brooks Crickard, Jianhua Fu, and Joseph C. Reese J. Biol. Chem., 291:9853-9870, 2016 |  |
| Quantification of Compactness and Local Order in the Ensemble of the Intrinsically Disordered Protein FCP1 Eric B. Gibbs and Scott A. Showalter J. Physical Chemistry B, 120:8960-8689, 2016 |  |
| Elimination of truncated recombinant protein expressed in Escherichia coli by removing cryptic translation initiation site Matthew J. Jennings, Adam F. Barrios and Song Tan Prot. Expr. and Purif., 12:17-21, 2016 |  |
| Multivalent Interactions by the Set8 Histone Methyltransferase With Its Nucleosome Substrate Taverekere S. Girish, Robert K. McGinty and Song Tan J. Mol. Biol, 428:1531-1543, 2016 |  |
| Preparation, Crystallization, and Structure Determination of Chromatin Enzyme/Nucleosome Complexes R.K. McGinty, R.D. Makde, S. Tan Methods in Enzymology, 573:43-65, 2016 |  |
2015
| Decoupling of divergent gene regulation by sequence-specific DNA binding factors Chao Yan, Daoyong Zhang, Juan Antonio Raygoza Garay, Michael M. Mwangi and Lu Bai Nucleic Acids Research, 43:7292-7305, 2015 |  |
| Molecular mechanisms of ribosomal protein gene coregulation Rohit Reja, Vinesh Vinayachandran, Sujana Ghosh, and B. Franklin Pugh Genes & Development, 29:1942-1954, 2015 |  |
| Ccr4-Not and TFIIS Function Cooperatively To Rescue Arrested RNA Polymerase II Arnob Dutta, Vinod Babbarwal, Jianhua Fu, Deborah Brunke-Reese, Diane M. Libert, Jonathan Willis and Joseph C. Reese Mol Cell Biol, 35:1915-1925, 2015 |  |
| Extranucleosomal DNA enhances the activity of the LSD1/CoREST histone demethylase complex Sang-Ah Kim, Nilanjana Chatterjee, Matthew J. Jennings, Blaine Bartholomew and Song Tan Nucleic Acids Research, 43:4868-4880, 2015 |  |
2014
| An Integrated Model of Multiple-Condition ChIP-Seq Data Reveals Predeterminants of Cdx2 Binding Shaun Mahony, Matthew D. Edwards, Esteban O. Mazzoni, Richard I. Sherwood, Akshay Kakumanu, Carolyn A. Morrison, Hynek Wichterle, David K. Gifford PLOS Computational Biology, 10:1-14, 2014 |  |
| Subnucleosomal Structures and Nucleosome Asymmetry across a Genome Ho Sung Rhee, Alain R. Bataille, Liye Zhang and B. Franklin Pugh Cell, 159:1377-1388, 2014 |  |
| Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome Robert K. McGinty, Ryan C. Henrici & Song Tan Nature, 514:591-596, 2014 |  |
