 | Molecular Biology |
| The core of CEGR research utilizes traditional and cutting edge biochemical and molecular biology methods. These include protein expression and purification systems, transcription and protein-DNA interaction assays, transgenic cells, genetics, CRISPR-Cas9, and anchor away depletions. | |
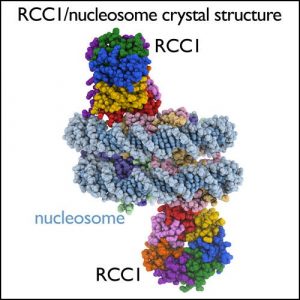 | Imaging |
| We use light microscopy, NMR, X-ray crystallography and electron microscopy imaging to visualize gene regulation molecules. For examining structures of molecules and interactions that contribute to gene regulation in solution, we use high resolution NMR spectrometers, equipped with cryogenic probe technology for signal enhancement. We grow crystals of chromatin complexes in temperature controlled incubators, and examine the diffraction properties of these crystals using our in-house X-ray crystallography facility. Penn State’s recent acquisition of an FEI Titan Krios microscope with Falcon III and Gatan K2 direct electron detectors provide us with state-of-the-art single particle cryoelectron microscopy capabilities. | |
 | DNA Sequencing |
| We were among the first in the world to apply deep sequencing to functional genomics. In 2007, we reported the first-ever ChIP-seq paper, which utilized the Applied Biosystems 454 sequencer to map the precise location of every nucleosome across the yeast genome. Since then we have utilized nearly every generation of deep sequencer including ABI SOLiD, and the Illumina GAII, HiSeq 2000, and NextSeq 500 series. | |
 | Bioinformatics |
| Many functional genomics assays are now based on deep sequencing, and thus produce vast amounts of data. Analyzing, integrating, and interpreting such data requires the development of novel computational methods. Uniquely, CEGR places computational biologists in the same laboratory space as molecular biologists. This enables close collaboration on the development of analysis methods for understanding gene regulation in high-resolution. | |
 | Mammalian Cell Culture Space |
| The shared mammalian cell culture space makes it possible for researchers to scale up for big experiments. The incubators can accomodate several liters of cells, which are protected by automatic CO2 tank switchovers. A pair of laminar flow hoods allows multiple labs to work at the same time. Finally, an EVOS XL Core digital microscope makes counting and imaging cells a breeze; a 60X long working distance fluorite objective is also available. |
